Advancing Microbiome-Based Biomarkers in Inflammatory Bowel Disease (IBD)
What key advancements could accelerate the implementation of microbiome biomarkers in clinical development?
Starting in February at the Microbiome Summit Barcelona 2025, @Eric Galvez delved into the challenges and opportunities in translating microbiome biomarkers into applications for Inflammatory Bowel Disease (IBD) clinical studies. The discussion addressed the critical role of the gut microbiome in IBD, characterized by compositional and functional dysbiosis, and its potential as a biomarker for diagnosis, activity monitoring and treatment response.
Overview of IBD and Microbiome Biomarkers
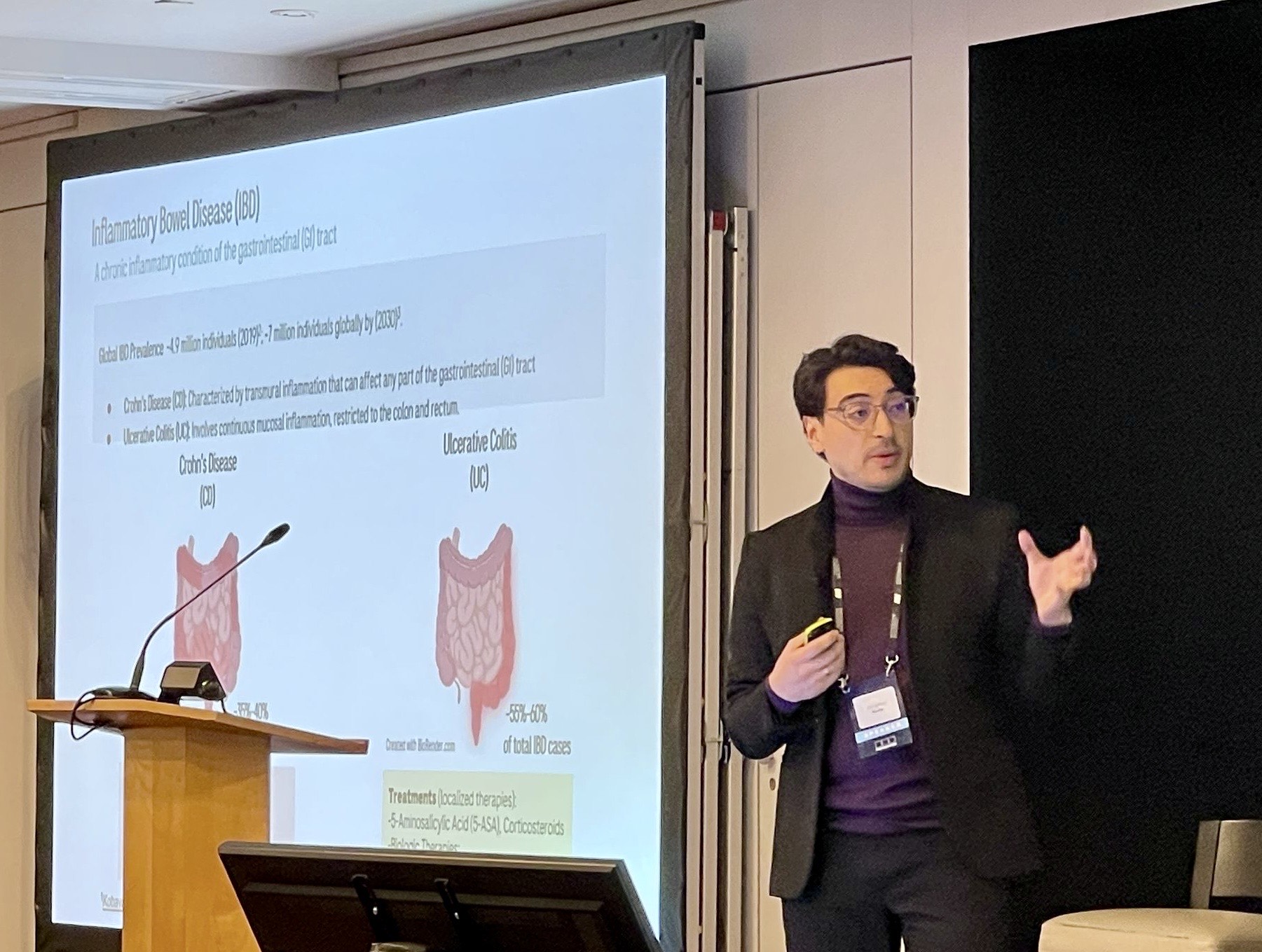
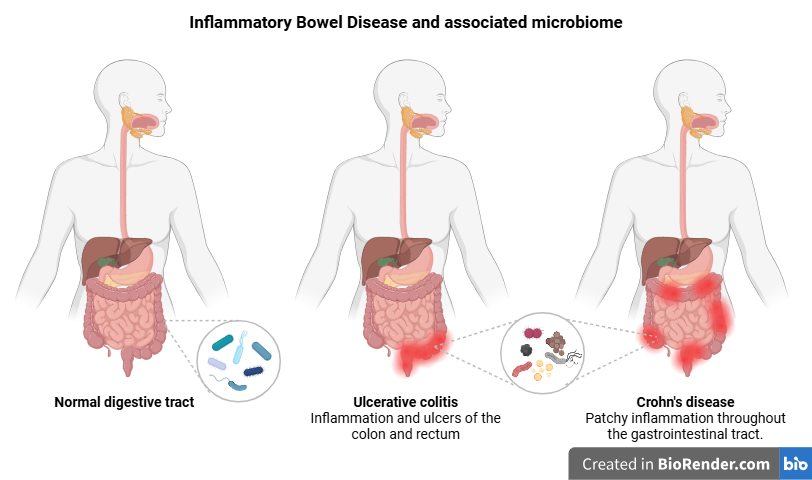
IBD is a chronic inflammatory condition of the gastrointestinal (GI) tract, significantly impacting patients’ quality of life. With an expected global prevalence of approximately 7 million by 2033 (Mak JWY, et al, MC Med Res Methodol. 2023), IBD primarily encompasses two main disease types; Crohn’s Disease (CD) and Ulcerative Colitis (UC). CD is marked by transmural inflammation that can affect any part of the GI tract, while UC involves continuous mucosal inflammation confined to the colon and rectum. Despite its increasing prevalence in industrialized countries, the cause of IBD is poorly understood due to its complexity and multifactorial nature, involving genetic, environmental, and microbiome factors.
Can we use gut metagenomic biomarkers for the Diagnosis and Prognosis of IBD to accelerate clinical development?
Toward the Development of a Noninvasive Diagnostic Tool
In a pivotal study recently published in Nature Medicine, Zheng et al. developed a microbiome-based diagnostic test using metagenomic data from fecal samples. This study reduced the complexity of the gut microbiome to just 10 specific bacterial species associated with IBD, achieving a high diagnostic accuracy (AUC > 0.90) across diverse populations. The authors translated their approach into a diagnostic test using the droplet digital PCR technique, a highly precise method for detecting tiny amounts of DNA or RNA from a liquid sample with a quick visual test, suggesting the potential of a multibacterial taxonomic biomarker panel as a noninvasive diagnostic tool for distinguishing active from non-active CD and UC samples vs healthy controls.
link to the original reference: DOI:10.1038/s41591-024-03280-4 (Journal: Nature Medicine, published: 04.10.2024)
Identification of Novel Host and Microbiome Biomarkers outperforming calprotectin
The recent study by Valdés-Mas et al., published in Cell, presents advancements in the use of microbiome biomarkers for diagnosing inflammatory bowel disease (IBD). Traditionally, calprotectin, a protein found in feces during inflammation, has been the primary biomarker for assessing IBD severity, albeit with moderate sensitivity and specificity. This study introduces a novel metagenome-informed metaproteomics (MIM) approach, which significantly enhances the diagnostic landscape for IBD.
Key Innovations:
- Improved Biomarker Identification: The MIM approach allows for the identification of functional disease-specific biomarkers by analyzing a curated protein database of prevalent host, microbiome, and dietary proteins using metagenomic sequences. This method could lead to the discovery of novel protein markers improving the diagnostic for microbiome-related diseases
- Reduction of Multidimensional Features: The study identified a pair of host-microbiome proteins out of millions of potential interactions that strongly correlate with IBD severity. This reduction prompts the interest in implementing novel protein assays that could outperform traditional biomarkers like calprotectin in diagnosing IBD and assessing disease severity.
- Dietary Influence on Microbiome: The research also explores how dietary shifts impact host-microbiome interactions, providing insights into the role of diet in IBD.
Overall, this research represents a significant step forward in the noninvasive diagnosis of IBD and its potential for disease monitoring and predicting treatment response using microbiome derived biomarkers.
Link to the original reference: DOI: 10.1016/j.cell.2024.12.016 (Journal: CELL, published: 01.20.2025)
Challenges and Opportunities
Implementing microbiome biomarkers in clinical settings requires standardization in sampling, sequencing, and data analysis techniques. However, advancements in multiomics integration, functional exploration, and replicability in large cohorts are expected to address these challenges.
Further validation in diverse and larger patient cohorts is necessary for integrating microbiome-based diagnostics into clinical practice. Prospective studies are needed to evaluate the real-world applicability and effectiveness of these biomarkers in predicting disease flares or potentially acts as endoscopy surrogate.
Conclusion
Advancements in microbiome-based biomarkers for IBD, particularly through noninvasive diagnostic tools and metaproteomics, hold promise for more precise and personalized approaches in IBD detection and management. Continued research and technological improvements are essential to overcome current challenges and fully realize the potential of these biomarkers in clinical development.
